COMPARISON OF METHODS FOR BULK AUTOMATED SIMULATION OF GLYCOSIDIC BOND CONFORMATIONS
1 N.D. Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Moscow, Russia
2 Chemical Faculty, National Research University Higher School of Economics, Moscow, Russia
KEYWORDS: carbohydrate, disaccharide, glycosidic bond conformation, molecular dynamics, NOE simulation, database filling, Nuclear Overhauser effect, force field
International Journal of Molecular Sciences, 2020, v.21(20), ID 7626 (pp. 1-19)
DOI: 10.3390/ijms21207626, PMID: 33076365
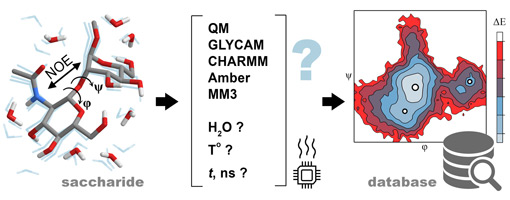
Six empirical force fields were tested for applicability to calculations for automated carbohydrate database filling. They were probed on eleven disaccharide molecules containing representative structural features from widespread classes of carbohydrates. The accuracy of each method was queried by predictions of nuclear Overhauser effects (NOEs) from conformational ensembles obtained from 50 to 100 ns molecular dynamics (MD) trajectories and their comparison to the published experimental data. Using various ranking schemes, it was concluded that explicit solvent MM3 MD yielded non-inferior NOE accuracy with newer GLYCAM-06, and ultimately PBE0-D3/def2-TZVP (Triple-Zeta Valence Polarized) Density Functional Theory (DFT) simulations. For seven of eleven molecules, at least one empirical force field with explicit solvent outperformed DFT in NOE prediction. The aggregate of characteristics (accuracy, speed, and compatibility) made MM3 dynamics with explicit solvent at 300 K the most favorable method for bulk generation of disaccharide conformation maps for massive database filling.