REStLESS: AUTOMATED TRANSLATION OF GLYCAN SEQUENCES FROM RESIDUE-BASED NOTATION TO SMILES AND ATOMIC COORDINATES
aKurnakov Institute of General and Inorganic Chemistry, Russian Academy of Sciences, Moscow, Russia
bZelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Moscow, Russia
KEYWORDS: SMILES, SMARTS, CSDB, glycan notation, atomic coordinates, structure visualization
Bioinformatics, 2018, v.34(15), pp. 2679-2681
DOI: 10.1093/bioinformatics/bty168, PMID: 29547883
Motivation
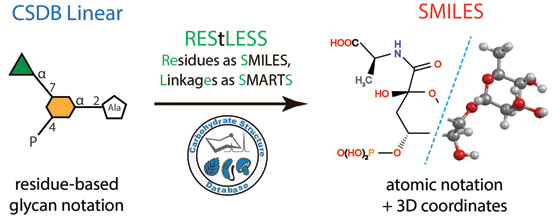 Glycans and glycoconjugates are usually recorded in dedicated databases in residue-based notations. Only a few of them can be converted into chemical (atom-based) formats highly demanded in conformational and biochemical studies. In this work, we present a tool for translation from a residue-based glycan notation to SMILES.
Glycans and glycoconjugates are usually recorded in dedicated databases in residue-based notations. Only a few of them can be converted into chemical (atom-based) formats highly demanded in conformational and biochemical studies. In this work, we present a tool for translation from a residue-based glycan notation to SMILES.
Results
The REStLESS algorithm for translation from the residue-based CSDB Linear notation to SMILES was developed. REStLESS stands for ResiduEs as SMILES and LinkagEs as SMARTS, where SMARTS reaction expressions are used to merge pre-encoded residues into a molecule. The implementation supports virtually all structural features reported in natural carbohydrates and glycoconjugates. The translator is equipped with a mechanism for the conversion of SMILES strings into optimized atomic coordinates. The obtained coordinates can be used as starting geometries for various computational tasks.
Availability
REStLESS is integrated in the Carbohydrate Structure Database (CSDB) and is also available both as a web-service and user web-tool at the CSDB platform (http://csdb.glycoscience.ru/database/core/translate.html#from).